Hi All
I am monthly-averaging daily ACCESS diagnostics and saving to files (1 sample per output file). Then I add to file time and time_bounds variables and relevant attributes. I can proceed to build a cosima dataset without any error messages, but using that dataset to load the actual variables fails with the following message:
ValueError: Could not find any dimension coordinates to use to order the datasets for concatenation.
The same error occurs whether I specify variable frequency or ncfile = '%monthly%' in the getvar function. I provide detail below, as well as in a notebook which can be found here:
/home/552/as2408/NotebookShare/DatabaseTest_V1.ipynb. If anyone wants to actually run the notebook parts where files are modified I can put clean netcdf files somewhere. Any advice would be appreciated.
This is how the averaging is done per file, pretty straightforward (with some obvious variable definitions):
ds = xr.open_dataset(FileNameIn);#Loading a file with 3 months of daily data
ds = ds.sel(time=time_slice) #Limiting to the desired month
Var=ds[varname].mean('time'); #choosing one variable of interest
Var.to_netcdf(FileNameOut) #saving to file
This is an example of how I add time variables to one file:
fldBinnedTest = '/g/data/g40/access-om2/archive/01deg_jra55v140_iaf_cycle3_antarctic_tracers/test/exptest/'
month=6; monthstr = str(month).zfill(2); year = 1981; yearstr = str(1981)
fndate = yearstr+'_'+monthstr
fn0 = 'passive_adelie-monthly-mean-ym_'+fndate+'.nc';#fn0 = 'passive_adelie-monthly-mean-ym_1981_04.nc'
from netCDF4 import Dataset
dsf = Dataset(fldBinnedTest+fn0, 'r+')#, format='NETCDF4_CLASSIC', diskless=True)
date1 = yearstr+'-'+str(month).zfill(2)+'-15'
print(date1);
t = pd.date_range(start=date1, end=date1, periods=1)
print(t)
from datetime import date
d0 = date(1900, 1, 1)
d1 = date(year, month, 15)
delta = d1 - d0
print(delta.days)
dsf.createDimension('time', None)
time = dsf.createVariable('time', np.int32, ('time',))
time[:] = delta.days
time.standard_name = 'time' # Optional
time.long_name = "time";
time.units = "days since 1900-01-01 00:00:00";
time.cartesian_axis = "T";
time.calendar_type = "GREGORIAN" ;
time.calendar = "GREGORIAN";
time.bounds = "time_bounds";
d1 = date(year, month, 1); delta = d1 - d0;
time_bound_1 = delta.days
if month<12:
d1 = date(year, month+1, 1); delta = d1 - d0;
else:
d1 = date(year+1, 1, 1); delta = d1 - d0;
time_bound_2 = delta.days
print(time_bound_1); print(time_bound_2)
dsf.createDimension('nv', 2)
time_bounds = dsf.createVariable('time_bounds', np.int32, ('time','nv'))
time_bounds[:] = [time_bound_1,time_bound_2]
time_bounds.standard_name = 'time_bounds'
time_bounds.long_name = 'time_bounds'
time_bounds.units = "days since 1900-01-01 00:00:00";
time_bounds.calendar = 'GREGORIAN'
dsf.close()
Finally, I build a dataset, e.g.,
sessionTestTime = cc.database.create_session(FoldDBs+'Exp9tracersTestTime.db')#
cc.database.build_index(fldBinnedTest,sessionTestTime)
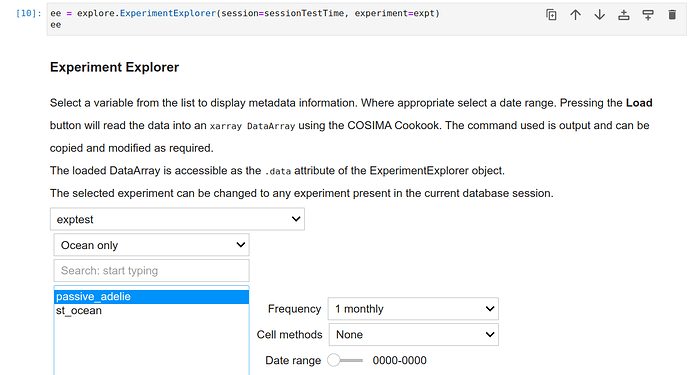
which ends without error and also shows up without error in the explorer, except the times (and cell_methods) are not identified:
And as I wrote above, getvar fails whether I use the following forms or some variations of them:
passive_adelie = cc.querying.getvar(expt='exptest', variable='passive_adelie',
session=sessionTestTime, frequency='1 monthly',
start_time='1980-01-01', end_time='1995-09-09')
passive_adelie = cc.querying.getvar(expt='exptest', variable='passive_adelie',
session=sessionTestTime, ncfile = '%monthly%')
e.g.,
---------------------------------------------------------------------------
ValueError Traceback (most recent call last)
Cell In[13], line 1
----> 1 passive_adelie = cc.querying.getvar(expt='exptest', variable='passive_adelie',
2 session=sessionTestTime, frequency='1 monthly')
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.10/lib/python3.9/site-packages/cosima_cookbook/querying.py:368, in getvar(expt, variable, session, ncfile, start_time, end_time, n, frequency, attrs, attrs_unique, return_dataset, **kwargs)
364 return d[variables]
366 ncfiles = list(str(f.NCFile.ncfile_path) for f in ncfiles)
--> 368 ds = xr.open_mfdataset(
369 ncfiles,
370 parallel=True,
371 combine="by_coords",
372 preprocess=_preprocess,
373 **xr_kwargs,
374 )
376 if return_dataset:
377 da = ds
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.10/lib/python3.9/site-packages/xarray/backends/api.py:1026, in open_mfdataset(paths, chunks, concat_dim, compat, preprocess, engine, data_vars, coords, combine, parallel, join, attrs_file, combine_attrs, **kwargs)
1013 combined = _nested_combine(
1014 datasets,
1015 concat_dims=concat_dim,
(...)
1021 combine_attrs=combine_attrs,
1022 )
1023 elif combine == "by_coords":
1024 # Redo ordering from coordinates, ignoring how they were ordered
1025 # previously
-> 1026 combined = combine_by_coords(
1027 datasets,
1028 compat=compat,
1029 data_vars=data_vars,
1030 coords=coords,
1031 join=join,
1032 combine_attrs=combine_attrs,
1033 )
1034 else:
1035 raise ValueError(
1036 "{} is an invalid option for the keyword argument"
1037 " ``combine``".format(combine)
1038 )
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.10/lib/python3.9/site-packages/xarray/core/combine.py:982, in combine_by_coords(data_objects, compat, data_vars, coords, fill_value, join, combine_attrs, datasets)
980 concatenated_grouped_by_data_vars = []
981 for vars, datasets_with_same_vars in grouped_by_vars:
--> 982 concatenated = _combine_single_variable_hypercube(
983 list(datasets_with_same_vars),
984 fill_value=fill_value,
985 data_vars=data_vars,
986 coords=coords,
987 compat=compat,
988 join=join,
989 combine_attrs=combine_attrs,
990 )
991 concatenated_grouped_by_data_vars.append(concatenated)
993 return merge(
994 concatenated_grouped_by_data_vars,
995 compat=compat,
(...)
998 combine_attrs=combine_attrs,
999 )
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.10/lib/python3.9/site-packages/xarray/core/combine.py:629, in _combine_single_variable_hypercube(datasets, fill_value, data_vars, coords, compat, join, combine_attrs)
623 if len(datasets) == 0:
624 raise ValueError(
625 "At least one Dataset is required to resolve variable names "
626 "for combined hypercube."
627 )
--> 629 combined_ids, concat_dims = _infer_concat_order_from_coords(list(datasets))
631 if fill_value is None:
632 # check that datasets form complete hypercube
633 _check_shape_tile_ids(combined_ids)
File /g/data/hh5/public/apps/miniconda3/envs/analysis3-22.10/lib/python3.9/site-packages/xarray/core/combine.py:149, in _infer_concat_order_from_coords(datasets)
144 tile_ids = [
145 tile_id + (position,) for tile_id, position in zip(tile_ids, order)
146 ]
148 if len(datasets) > 1 and not concat_dims:
--> 149 raise ValueError(
150 "Could not find any dimension coordinates to use to "
151 "order the datasets for concatenation"
152 )
154 combined_ids = dict(zip(tile_ids, datasets))
156 return combined_ids, concat_dims
ValueError: Could not find any dimension coordinates to use to order the datasets for concatenation