Hi @ongqingyee,
My understanding is that fld_s08i234: surface_runoff_flux comes from the land model, and refers to surface runoff over the land points rather than the runoff going into the ocean. If you have access to it, the variable fld_s26i004: water_flux_into_sea_water_from_rivers should contain the river runoff going into the ocean from the UM.
If the runoff from the UM hasn’t been saved, you might be able to take it from the ocean output. The runoff variable saved by MOM will contain the same data, but regridded onto the ocean grid.
Based on the coupling variables sent to the ocean in ESM1.5 here I don’t think there was any equivalent to licalvf in ESM1.5 (it’s being added in 1.6!). I believe all the freshwater flux from the ice sheets was distributed into the liquid runoff around the coastlines.
I followed the instructions here and ended up with this error, which I traced back to here.
I am slightly confused by how the MOM5 ESM1.5 component code is showing up when running ACCESS-OM2 with ESM ocean restarts.
There are a few different versions of the MOM5 code in different places, and the latest release of OM2 should be using the code from this repository. I think this error is coming from here in the FMS library used by OM2.
The MOM5 restarts contain checksums of each variable’s data, e.g:
double temp(Time, zaxis_1, yaxis_1, xaxis_1) ;
temp:long_name = "temp" ;
temp:units = "none" ;
temp:checksum = "DCA837A688447387" ;
When starting from a restart, the model reads in the restart variables and recalculates the checksums. It then compares these to the checksums stored in the restart file, exiting with an error if they don’t match.
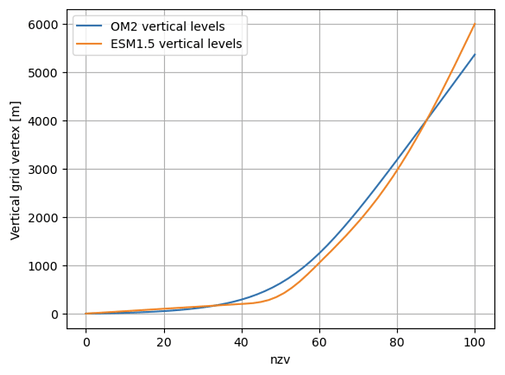
Based on the earlier discussion, did you end up vertically interpolating the ESM1.5 restarts? As this would change the restart data, it would cause this check to fail. It looks like you can add
checksum_required=.false.
to ocean/input.nml in the fms_io_nml section to switch off this check