Ok, so it looks like analysis3 uses notebook==7.4.5 and jupyterlab==4.4.6.
Neither of those are a major version out from the suggested fix, so I’ll try rebuilding the environment with those and see what happens. I’ll also check if we can solve the environment with the default ARE versions to see if that ameliorates the issue - Jemma’s post indicates to me it might - but I’m less confident we’ll be able to do that.
Background on the issue
This is almost certainly not a python issue, and is probably a javascript issue instead. For anyone who’s ever wondered why you get weird git diffs like this when you look at notebooks:
index 408101d..0ca38d5 100644
--- a/Untitled-1.ipynb
+++ b/Untitled-1.ipynb
@@ -2,7 +2,7 @@
"cells": [
{
"cell_type": "code",
- "execution_count": 1,
+ "execution_count": 2,
"id": "25423f80",
"metadata": {},
"outputs": [
@@ -24,7 +24,7 @@
" 86, 87, 88, 89, 90, 91, 92, 93, 94, 95, 96, 97, 98, 99])"
]
},
- "execution_count": 1,
+ "execution_count": 2,
"metadata": {},
"output_type": "execute_result"
}
@@ -39,6 +39,38 @@
"\n",
"np.arange(1,100)"
]
+ },
+ {
+ "cell_type": "code",
+ "execution_count": 3,
+ "id": "2de0c1bf",
+ "metadata": {},
+ "outputs": [
+ {
+ "data": {
+ "text/plain": [
+ "[<matplotlib.lines.Line2D at 0x1485012e0>]"
+ ]
+ },
+ "execution_count": 3,
+ "metadata": {},
+ "output_type": "execute_result"
+ },
+ {
+ "data": {
+ "image/png": "iVBORw0KGgoAAA..nP8PLQdhZMGa0AAAAAAASUVORK5CYII=",
+ "text/plain": [
+ "<Figure size 640x480 with 1 Axes>"
+ ]
+ },
+ "metadata": {},
+ "output_type": "display_data"
+ }
+ ],
+ "source": [
+ "import matplotlib.pyplot as plt\n",
+ "plt.plot(np.arange(1,100))"
+ ]
}
],
"metadata": {
It’s because a jupyter notebook is actually a .json file in a trenchcoat. json (Javascript object notation) is what a server and a Single Page Application (SPA, think anything built in react, ie. Facebook, Instagram, Xero, etc etc) use to communicate. It’s also how anything built using Electron (ie. VS Code, Teams, Balena Etcher) works. The Jupyter frontend apparently uses react with some custom extra bits - I’ll come back to that.
Basically, when you have an interactive web application (ie. a jupyter notebook), and a running server - jupyterlab does this - here is an excerpt from an ARE session PBS log:
[I 2025-09-30 15:30:49.162 ServerApp] Serving notebooks from local directory: /home/189/ct1163/.jupyter-root
[I 2025-09-30 15:30:49.162 ServerApp] Jupyter Server 2.16.0 is running at:
[I 2025-09-30 15:30:49.162 ServerApp] http://localhost:42996/node/gadi-cpu-bdw-0019.gadi.nci.org.au/42996/lab
[I 2025-09-30 15:30:49.162 ServerApp] http://127.0.0.1:42996/node/gadi-cpu-bdw-0019.gadi.nci.org.au/42996/lab
[I 2025-09-30 15:30:49.162 ServerApp] Use Control-C to stop this server and shut down all kernels (twice to skip confirmation).
[W 2025-09-30 15:30:49.170 ServerApp] No web browser found: Error('could not locate runnable browser').
Discovered Jupyter Notebook server listening on port 42996!
they will communicate by sending blobs of json back and forth.
So where does it all go wrong?
TLDR; Javascript is a (glorious) mess, and it’s very easy to shoot yourself in the foot with it. I don’t know exactly why or where the issue is coming from, but because of the necessarily asynchronous way that web apps are designed, it’s very easy to cause subtle bugs with race conditions, etc, which lead to issues like these.
Race conditions are almost certainly the source of the problems here - when you send a really large output - think a massive plot, output ten million lines of warnings, or just print a gigantic xarray dataarray, the jupyterlab server has to send all that data as a json blob back to the client (web browser javascript engine). This takes a while. A few years ago, I tried to send an entire pandas dataframe (~100k rows) as a json blob, and it took about a minute, from a server running locally on my machine to the browser on my machine.
Why is this a problem? Because in the meantime, whilst your json blobs are sent over the wire, you can send other instructions to the browser. Remember how I said jupyter uses react? React uses a thing called a Virtual DOM (Document Object Model - it’s basically a javascript object that represents the web page you have open), and that does it’s best not to render everything at once. When stuff scrolls out of view, react unmounts it. If stuff is still being sent over the wire at this point & the author of the jupyterlab code handling this wasn’t very careful, all manner of weird bugs can occur.
N.B: The client and server will be firing JSON back and forth continuously - not just when you execute a bit of code. You can create your own footguns, but the browser has plenty of it’s own.
With all that in mind, some things you can do to hopefully reduce the issue, whilst we see if we can find a version of jupterlab without this particular bug:
- Don’t keep too much output hanging around, if possible. Notebooks that look like this
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
warnings.warn("This is a warning")
/var/folders/qg/vhg6q9gn4zl01xbg1mfw6jzhxzf1wt/T/ipykernel_10498/1352830306.py:5: UserWarning: This is a warning
are going to be particularly prone to it. Clearing the duplicated warnings is probably going to improve the situation, at least a bit.
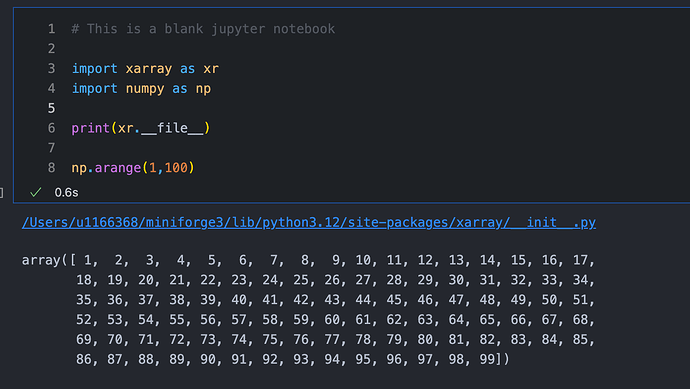
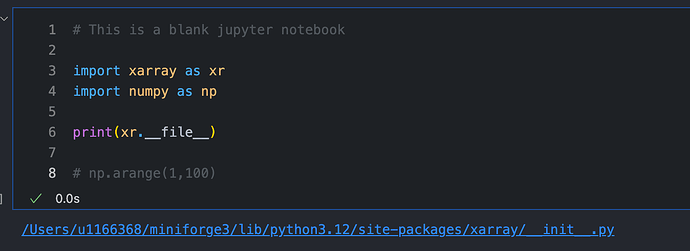
2. Don’t print out massive arrays constantly in order to look at them. Stuff like this:
is likely also going to cause issues. Unassigned statements at the end of code cells in order to view the outputs should be cleaned up when you’re done looking at them - ie. change it to this:
Really, it boils down to being mindful of the amount of data that you’re sending back and forth. If it gets too big, the issue is likely to occur - especially if you start triggering lots of other browser events.
Anyhoo, I’ll see if we can update those package versions & whether that helps at all.